MacSyFinder macromolecular system visualization
What is MacSyFinder ?
Learn more about MacSyFinder.
Reference:
How to explore a MacSyFinder macromolecular system ?
You can access to the MacSyFinder macromolecular system visualization window by clicking on the label indicated in the Macromolecular system field in the Macromolecular Systems table.
This window allows you to access to a detailled description of the selected macromolecular system components.
You can also use the main navigation menu in the Comparative Genomics section and Macromolecular Systems subsection to obtain the MacSyFinder predictions page. This page enumerates all macromolecular systems detected for the selected organism and its replicons.
What is the ‘Genomic Objects’ table ?
The table Genomic Objects provides informations regarding the genomic objects present in the macromolecular system.
You can export the genes by clicking on Export to Gene Cart.
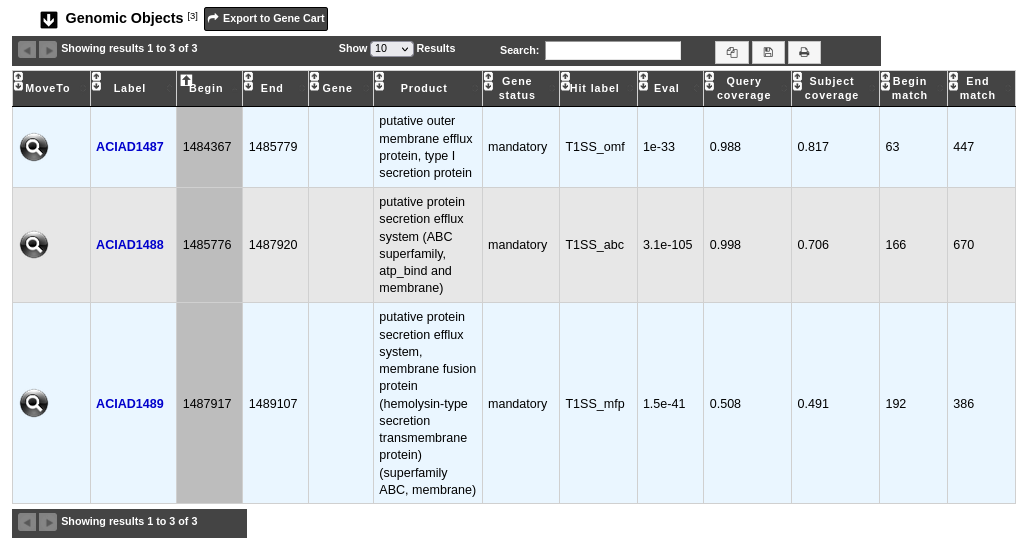
Label: Label of the genomic object. Click on it allow to access to its annotation page.
Begin and End: Location of the genomic object on the sequence.
Gene: Gene name if any.
Product: Description of the gene product of the genomic object.
Gene status: Status of the gene in the system (mandatory, accessory, neutral).
Hit label: Name of the MacSyFinder HMM profile which matchs with the sequence.
Eval: Evalue of the match.
Query coverage: Coverage of the match on the sequence.
Subject coverage: Coverage of the match on the MacSyFinder HMM profile.
Begin match and End match: Location of the match on the sequence.