Secondary metabolites¶
What are secondary metabolites?¶
Secondary metabolism (also called specialized metabolism) is a term for pathways and small molecule products of metabolism that are not absolutely required for the survival of the organism. Secondary metabolites are produced by many microbes, plants, fungi and animals. Bacterial secondary metabolites are an important source of antimicrobial and cytostatic drugs. These molecules are often synthesized in a stepwise fashion by multimodular megaenzymes that are encoded in clusters of genes encoding enzymes for precursor supply and modification.
What is antiSMASH?¶
Antismash is a tool predicting secondary metabolite gene clusters in bacterial genomes.
These result are linked to The Minimum Information about a Biosynthetic Gene cluster (MIBiG) database.
How to access to the secondary metabolites gene clusters predicted by antiSMASH?¶
Secondary metabolites gene clusters predictions are available through the Metabolism section, in the main navigation menu.
What is the “Predicted secondary metabolite clusters” table?¶
This table enumerates all secondary metabolite clusters predicted for the selected organism and its replicons. Each predicted cluster is associated to a Cluster type defined by antiSMASH.
- Region type region type predicted by antiSMASH
- MIBiG link to MIBiG best hit (if any)
- Completion completion of the best hit between MIBiG region and antiSMASH prediction region
- Product product of the MIBiG compound
- Type type of the MIBiG compound
MIBiG completion¶
The completion is computed as follow :
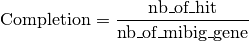
Where:
= number of genes with blast hit in the antiSMASH predicted region and MIBiG region
= number of MIBiG genes (all of them) in the MIBIG curated region
Meaning that when 2 or more genes in a single MIBiG curated region are similar, the same gene in pkgdb can hit on these MIBiG gene. When that happen, the completion can be higher than 1 (represented by 1* or the real number).
How to explore a secondary metabolite cluster?¶
The AntiSMASH cluster visualization window can be accessed by clicking on any cluster number in the Cluster field. This window allows you to visualize the full antiSMASH cluster prediction and its genomic context.