Mapping Overview (Variant/RNA-Seq)
Overviewing RNA-Seq or Variant profiling experiments results
This page allows users to have a complete summary of the mapping process for each experiment that have been performed on the studied organism. Results are reported in tables that can be easily expanded/collapsed by clicking on the small horizontal arrow.
An Example is given below in the case of Helicobacter Pylori public data :
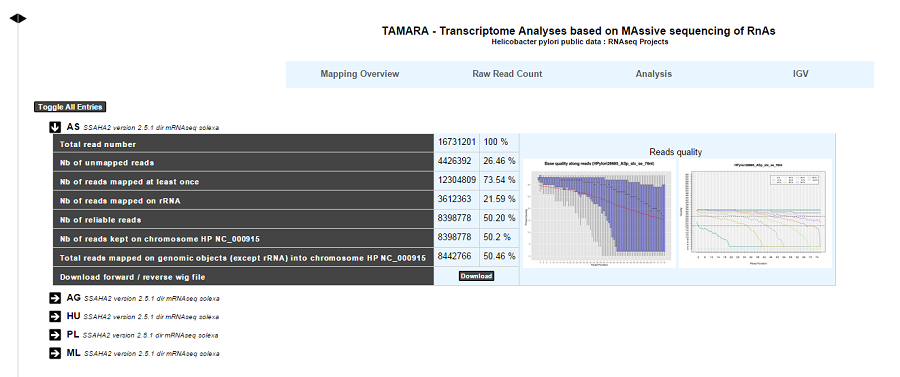
For each experiment, user will have access to the following data:
The total read number;
The number of unmapped reads;
The number of reads mapped at least once;
The number of reads that matched rDNA : Each mapped read is not count once but 1/(number of times mapped on genome);
The number of reliable reads (with mapping quality values not null);
Nb of reads kept on … : Number of mapped reads against a specific chromosome or plasmid;
Total reads mapped on genomic objects (except rRNA) into … : Number of mapped reads except rRNA.