EGGNOG Automatic Classification based on COG categories
eggNOG database (version 5.0.2)
eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) is a public resource in which thousands of genomes are analyzed at once to establish orthology relationships between all their genes.
eggNOG focuses on providing: (i) comprehensive functional annotations for the inferred orthologs, (ii) predictions across thousands of genomes covering the three domains of life and viruses, and iii) hierarchical resolution of orthology assignments and fine-grained relationships (i.e. in-paralogies) based on phylogenetic analysis.
An Orthologous Group (OG) is defined as a cluster of three or more homologous sequences that diverge from the same speciation event.
Orthologous Groups were functionally annotated using updated versions of Gene Ontology, KEGG pathways, SMART/PFAM domains and expanded to CAZy and KEGG modules. Moreover, general free text descriptions and COG functional categories were updated for each OG.
More information about the method: http://eggnogdb.embl.de/#/app/methods
eggNOG-mapper (version 2.1.12)
eggNOG-mapper is a tool for fast functional annotation of novel sequences. It uses precomputed orthologous groups and phylogenies from the eggNOG database to transfer functional information from fine-grained orthologs only. Common uses of eggNOG-mapper include the annotation of novel genomes, transcriptomes or even metagenomic gene catalogs.
The use of orthology predictions for functional annotation permits a higher precision than traditional homology searches (i.e. BLAST searches), as it avoids transferring annotations from close paralogs (duplicate genes with a higher chance of being involved in functional divergence).
We run eggnog-mapper using EGGNOGDB and diamond for the alignement.
More information about eggNOG-mapper: http://eggnog-mapper.embl.de/
EGGNOG Automatic Classification page
This page presents statistics on reference genome genes classified by eggNOG in Orthologous Groups (OG) by COG functional categories.
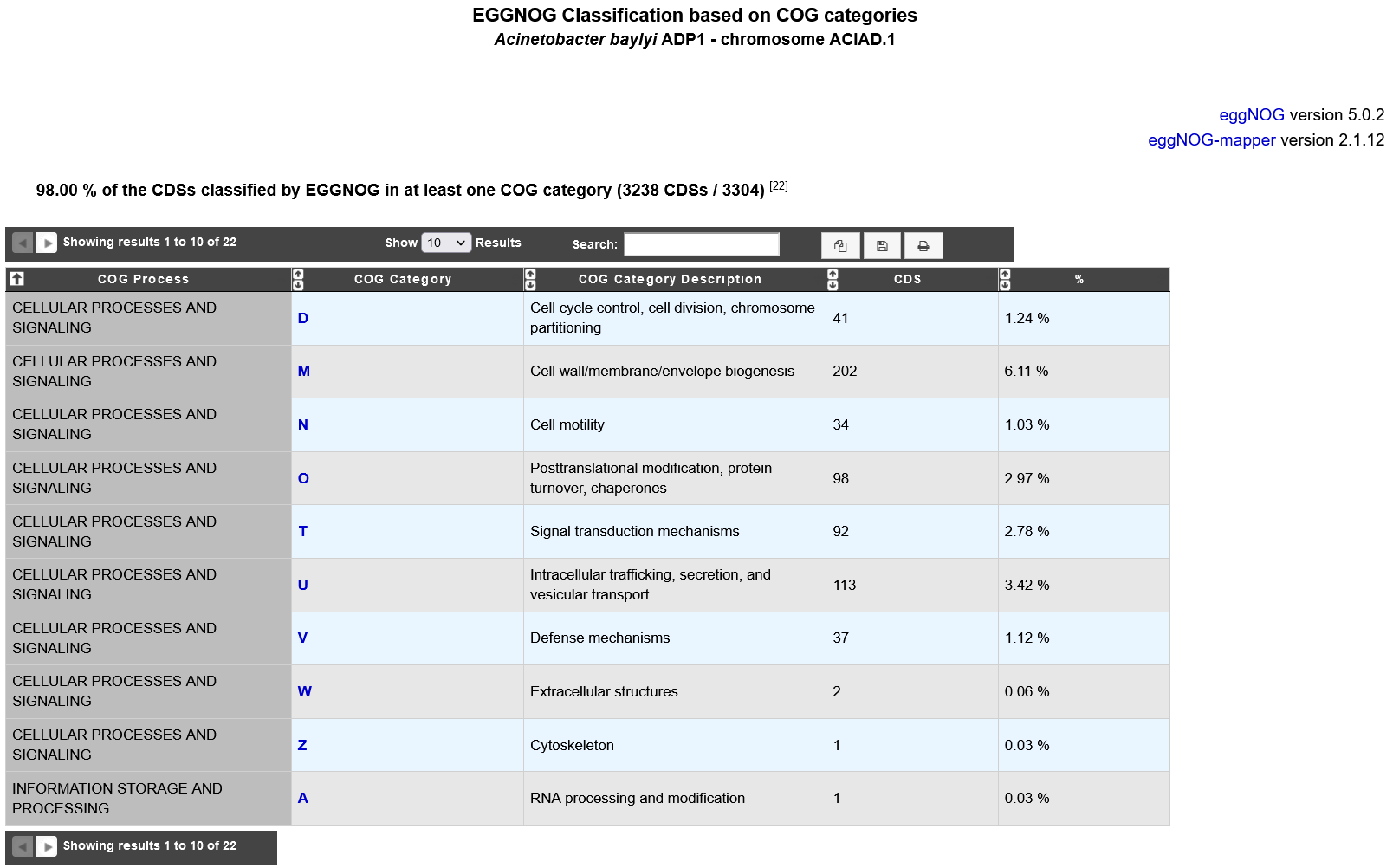
COG Process: COG functional process name.
COG Category: COG functional category ID.
COG Category Description: COG functional category description.
CDS: Number of the CDSs classified by eggNOG in at least one COG category.
%: Percentage of the CDSs classified by eggNOG in at least one COG category.
Clicking on a COG category displays all genes assigned to OG in that COG functional category.
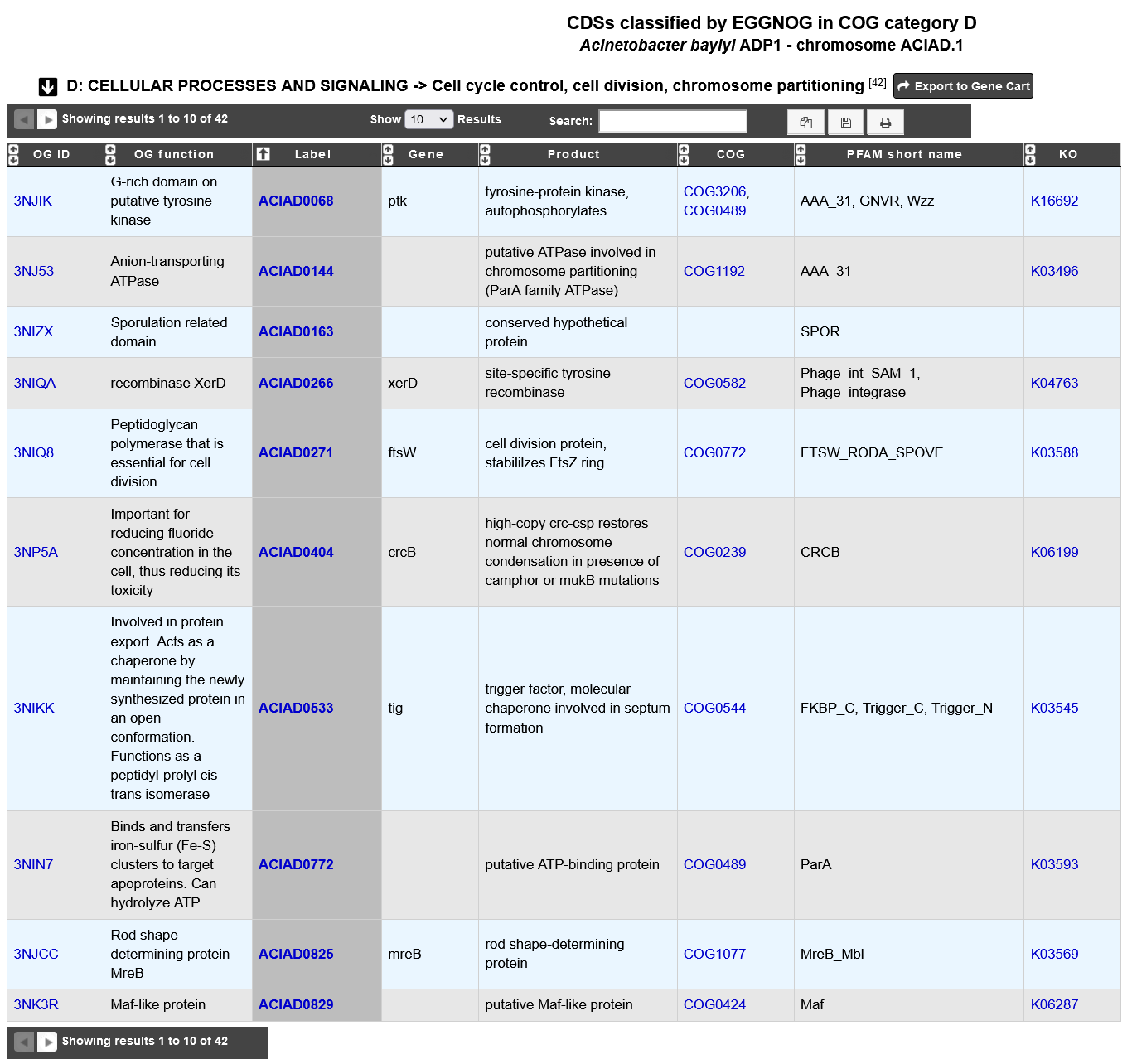
OG ID: eggNOG Orthologous Group id.
OG function: eggNOG Orthologous Group functional description.
Label: Label of the protein. If you click on the label, you access to the Gene annotation window.
Gene: Gene name of the protein if any.
Product: Product description of the protein.
COG: Clusters of Orthologous Group name (COG).
PFAM short name: InterPro PFAM entry name.
KO: KEGG Ortholog entry ID.