RNAseq Read Count Analysis¶
Analyzing Read Count¶
According to this tool, it is possible to know exactly how many reads matched a given genomic object of the reference sequence. Results are accessible following a 5 steps process which is described below.
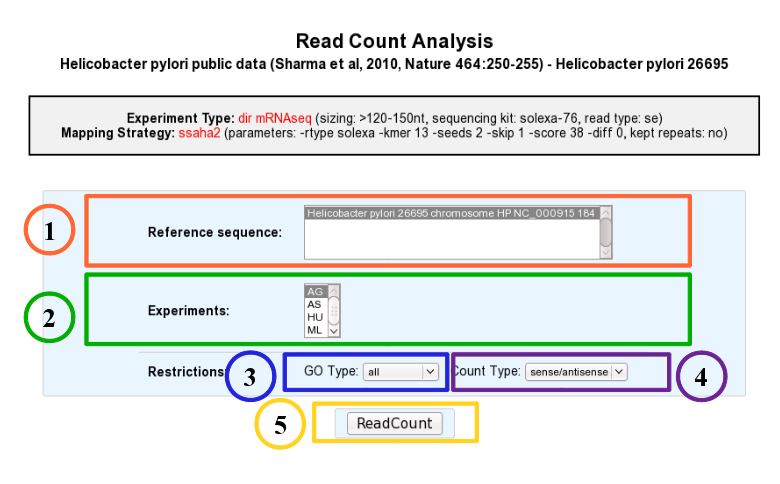
- 1. Choose one or several reference sequences.
- 2. Select at least one experiment and compute the associated read count number per genomic object. (check publication for terminology of experiments, which is displayed in the head of the interface: Sharma et al, 2010, Nature 464:250-255 for the given example)
- 3. It is possible to restrict the query to one or several given classes of genomic objects ( CDS, fCDS, rRNA, tRNA, miscRNA or all ).
- 4. Query can be constrained upon the strand of the transcripts (direct, reverse, both)
- 5. Submit query.
As usual, results are reported in a table which is composed of 3 main sections (see below).
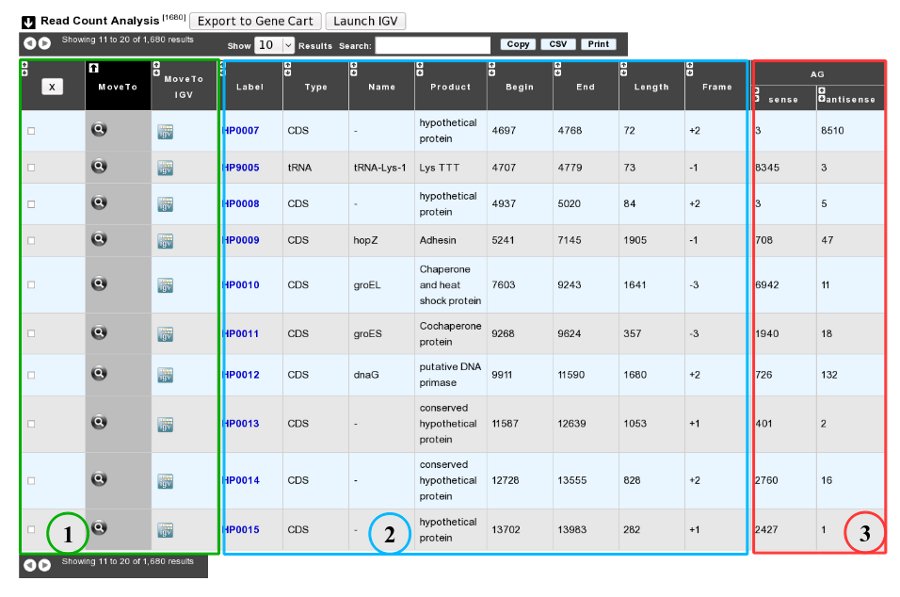
1. Export functions. This section allows users to make all genes (or subsets of genes) available for other analysis tools. 3 main operations are possible here:
- select subsets of genes (by selecting checkboxes on the first column) and export them into a Gene Cart by using the “Export To Gene Cart” button.
- See one selected gene into the MaGe Genome Browser by clicking on the magnifying glass.
- Direct link to the selected gene in Integrative Genome Viewer.
2. The second part reports the main genomic object features : Label (Link to more Genomic Object information), Type, Name, Product, Begin, End, Length, Frame.
3. RNA-Seq Result part : Read count (direct and/or reverse)