Export Data¶
This page allows to export data from MicroScope in several formats. The page has 2 modes. They are presented below with their export possibilities.
Replicon mode¶
Here is an overview of this mode:
This tool allows to retrieve data stored in PkGDB from a specific sequence. This information can be downloaded in the most common file formats (EMBL, GenBank, Fasta, GFF, tab delimited). Moreover, data on role categories used in MicroScope, and/or MicroCyc metabolic Pathway/Genome database (PGDBs) can be downloaded too. See below for a more precise presentation.
To use this mode, first select a sequence and a reference replicon with the Simple Selector at the top right corner of the interface. After that, you can use the various export possibilities described below.
Extract genome¶
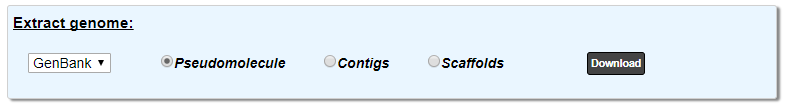
This part allows you to download the sequence in various formats. You can choose to extract:
the pseudomolecule (concatenated contigs or complete sequence if the genome is finished)
the contigs
the scaffolds
Use the drop down list to choose the format among:
Extract data¶
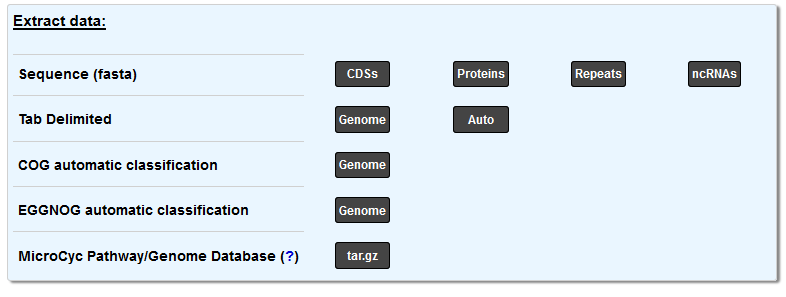
The first line of buttons allows to download several parts of the sequence (in the FASTA format):
CDSs will download the nucleic sequence of all the CDS of the sequence
Proteins will download the protein sequence of all the CDS of the sequence
Repeats will download the nucleic sequence of the repeated regions of the sequence
ncRNAs will download the nucleic sequence of all the non-coding RNAs of the sequence
The second line allows to extract the annotations (in a tab delimited format):
Genome will download the current annotation of all genomic objects in the sequence
Auto will download the automatic annotation of all genomic objects in the sequence
The Genome button on the fourth line allows to download the EGGNOG Automatic Classification.
Finally, you can download the MicroCyc Pathway/Genome Database of the selected genome with the last button.
Extract a region¶
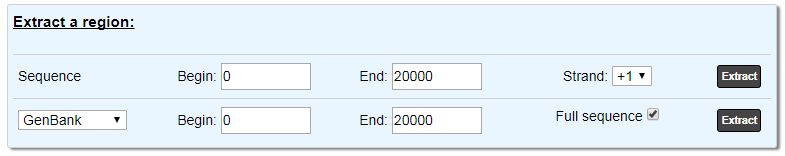
The first line allows you to extract a region of the nucleic sequence: select the Begin, End positions and the strand you want to get. The default values correspond to the current region of the Genome Browser. Click on Extract to retrieve the results. The data are exported in the FASTA format.
The second part allow you to extract annotations of the objects in the region indicated by Begin and End:
With the Full Sequence option, you will obtain the annotation of the objects in the region with the coordinates on the full sequence.
If this option is disabled, you will obtain the annotation within the region but the location will be computed relatively to the region (in this case, only the sequence of the region is included).
Use the drop down list to choose the format.
Noncoding DNA¶

This part allows to extract the ncDNA sequences from a sequence. Indicate a minimal length and include, if necessary, the RNAs. Click Extract to retrieve the data.
Extract a sequence fragment¶
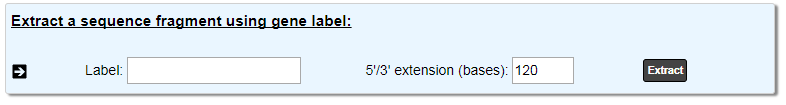
This tool allows to extract a sequence fragment around a genomic object by indicating its label and the 5’/3’ extension length.
Extract Classification¶
The Role Classification button allows to get the complete Role Classification in a text format.
The BioProcess Classification button allows to get the complete BioProcess Classification in a text format.
Organism mode¶
Here is an overview of this mode:
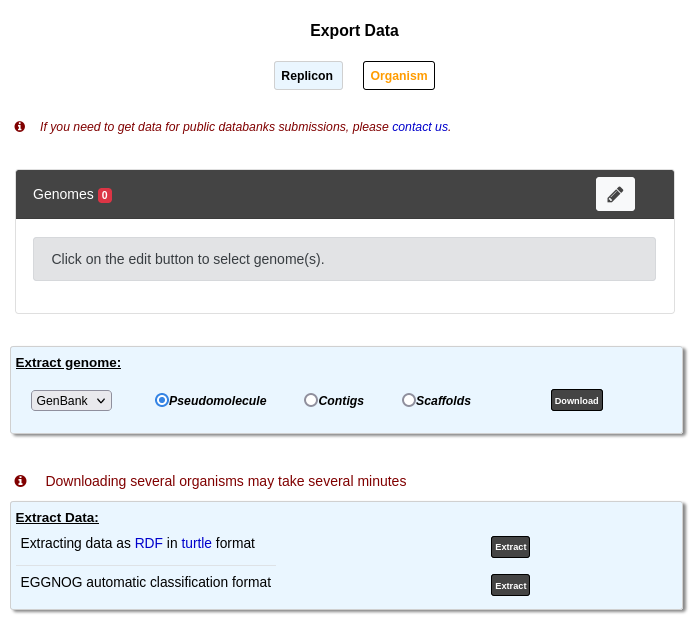
This tool allows to retrieve data stored in PkGDB from a group of genomes. Extraction of several genomes may take several minutes. This page uses the Advanced Selector to select the genomes to export.
The Extract Genome and EGGNOG parts are similar to the Replicon mode except of course they retrieve information for all the selected genomes.
Therefore, we will only describe the Export Data as RDF part.
Export Data as RDF¶
This tool is used to export data in RDF to load it for example in a SPARQL triplestore. The RDF file format used by the MicroScope platform is the Turtle format.
SPARQL Request examples¶
Prefixes¶
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX mso: <https://mage.genoscope.cns.fr/microscope/ontology/#>
PREFIX mage: <https://mage.genoscope.cns.fr/microscope/mage/info.php?id=>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX obo: <http://purl.obolibrary.org/obo/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX faldo: <http://biohackathon.org/resource/faldo#>
PREFIX up_core: <http://purl.uniprot.org/core/>
PREFIX ec: <http://purl.uniprot.org/enzyme/>
PREFIX ncbi_tax: <https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=>
PREFIX rh: <http://rdf.rhea-db.org/>
PREFIX metacyc: <https://metacyc.org/META/NEW-IMAGE?type=NIL&object=>
Requests¶
# All genes of an organism from its taxID
# Organism: Acinetobacter sp. ADP1
# Taxonomy ID: 62977
SELECT DISTINCT ?genes WHERE {
?genes rdf:type obo:SO_0000704 ;
obo:RO_0002162 ?org .
?org mso:taxon ncbi_tax:62977 .
}
# All proteins of an organism from its taxID
# Organism: Acinetobacter sp. ADP1
# Taxonomy ID: 62977
SELECT DISTINCT ?protein WHERE {
?transcript obo:SO_transcribed_from ?genes ;
obo:SO_translate_to ?protein .
?genes rdf:type obo:SO_0000704 ;
obo:RO_0002162 ?org .
?org mso:taxon ncbi_tax:62977 .
}
# All genes (and nucleic sequence), proteins (and amino acid sequence)
# of an organism from its taxID
# Organism: Acinetobacter sp. ADP1
# Taxonomy ID: 62977
SELECT DISTINCT ?genes ?protein ?desc ?nucSeq ?protSeq WHERE {
?genes rdf:type obo:SO_0000704 ;
mso:hasSequence ?nucSeqObj ;
obo:RO_0002162 ?org .
?org mso:taxon ncbi_tax:62977 .
?nucSeqObj rdfs:value ?nucSeq .
?transcript obo:SO_transcribed_from ?genes ;
obo:SO_translate_to ?protein .
?protein a mso:Protein ;
dc:description ?desc ;
mso:hasSequence ?protSeqObj .
?protSeqObj rdfs:value ?protSeq .
}
# Get Gene-Protein-Reaction (GPR) associations
# of an organism from its taxID
# Organism: Acinetobacter sp. ADP1
# Taxonomy ID: 62977
SELECT DISTINCT ?genes ?protein ?reaction WHERE {
?transcript obo:SO_transcribed_from ?genes ;
obo:SO_translate_to ?protein .
?genes rdf:type obo:SO_0000704 ;
obo:RO_0002162 ?org .
?org mso:taxon ncbi_tax:62977 .
?reaction mso:isCatalyzedBy ?protein .
}