Macromolecular Systems¶
What type of Macromolecular systems can be detected?¶
a broad range of secretion systems: T1SS, T2SS,T3SS,T4SS, T5SS, T6SS, T9SS, Flg, T4P, Tad (Abby SS et al., Sci. Rep. 2016)
CRISPR-Cas systems: Clustered regularly interspaced short palindromic repeats (CRISPR) arrays and their associated Cas (CRISPR-associated) proteins form the CRISPR-Cas system. CRISPR-Cas are sophisticated adaptive immune systems that rely on small RNAs for sequence-specific targeting of foreign nucleic acids such as viruses and plasmids.
What is MacSyFinder?¶
Macromolecular System Finder (MacSyFinder) provides a flexible framework to model the properties of molecular systems (cellular machinery or pathway) including their components, evolutionary associations with other systems and genetic architecture. Modelled features also include functional analogs, and the multiple uses of a same component by different systems. Models are used to search for molecular systems in complete genomes or in unstructured data like metagenomes. The components of the systems are searched by sequence similarity using Hidden Markov model (HMM) protein profiles. The assignment of hits to a given system is decided based on compliance with the content and organization of the system model.
Learn more about MacSyFinder here.
Reference:
What is CRISPRCasFinder?¶
CRISPRCasFinder is a tool that allows to identify CRISPR arrays and Cas proteins. The CRISPR detection is based on Vmatch (a software for large scale sequence analysis) which identify all regularly-interspaced repeated sequences. CRISPRCasFinder associates an evidence level with each CRISPR detected using 3 criteria:
An entropy-based conservation index of repeats (EBcon);
The number of spacers ;
The overall percentage identity of spacers.
More information about CRISPRCasFinder see https://crisprcas.i2bc.paris-saclay.fr/.
Note
In MicroScope, CRISPRCasFinder is used only to detect CRISPR systems. Cas systems are detected by MacSyFinder.
References:
Abouelhoda et al. 2004. Replacing suffix trees with enhanced suffix arrays. J. Discrete Algorithms.
How to access to MacSyFinder and CRISPRCasFinder predictions?¶
MacSyFinder and CRISPRCasFinder predictions are available through the Comparative Genomics section, in the main navigation menu.
What is the ‘Macromolecular Systems’ table?¶
This table enumerates all macromolecular systems predicted for the selected organism and its replicons.
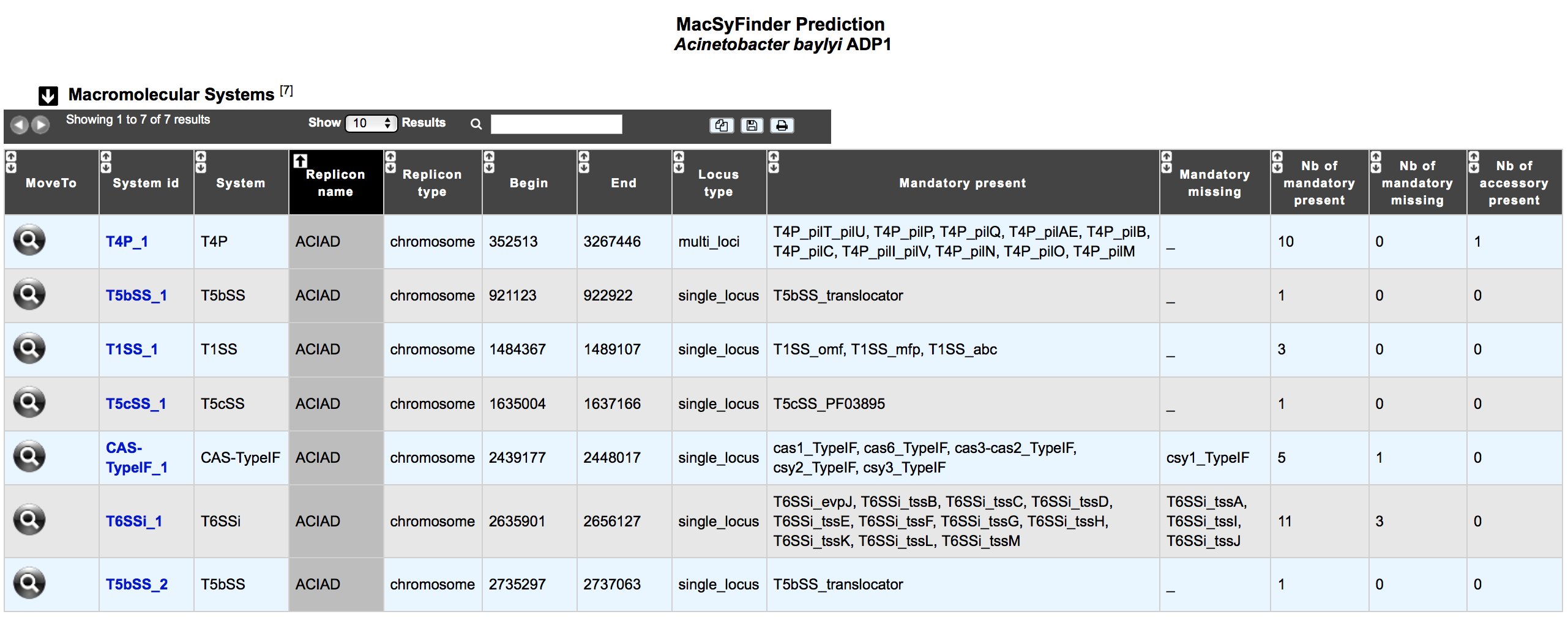
System id: identifier of the system in the organism
System: type of system detected by MacSyFinder
Replicon name: identification of the replicon
Replicon type: chromosome, plasmid or WGS
Begin / End: position of the system on the replicon
Locus type: single or multi locus
Mandatory present: list of mandatory genes of the system identified in the organism
Mandatory missing: list of mandatory genes of the system not detected in the organism
Nb of mandatory present: number of mandatory genes of the system identified in the organism
Nb of mandatory missing: number of mandatory genes of the system not detected in the organism
Nb of accessory present: number of accessory genes of the system identified in the organism
What is the ‘CRISPR’ table?¶
This table displays all CRISPR detected by CRISPRCasFinder and all Cas detected by MacSyFinder.
System id: identifier of the system in the organism
System: type of system detected (CRISPR or Cas)
Replicon name: identification of the replicon
Replicon type: chromosome, plasmid or WGS
Begin / End: position of the system on the replicon
Nb spacers / genes: number of CRISPR spacers / Number of Cas genes
Consensus repeat / Present gene: consensus repeat sequence predicted by CRISPRCasFinder / list of mandatory Cas genes
Evidence level: evidence level as computed by CRISPRCasFinder
How to explore a Macromolecular System?¶
The MacSyFinder System visualization window can be accessed by clicking on any cluster number in the System id field. This window allows you to access to a detailled description of a selected Macromolecular System.