IntegronFinder cluster visualization
What is IntegronFinder ?
Know more about IntegronFinder.
Reference:
How to explore an Integron cluster ?
You can access to the IntegronFinder cluster visualization window by clicking on the number indicated in the Integron id field in the Integron clusters table.
This window allows you to access to a detailled description of the integron structure.
You can also use the main navigation menu in the Comparative Genomics section and Integrons subsection to obtain the IntegronFinder predictions page. This page enumerates all integrons detected for the selected organism and its replicons.
What is the ‘Integron Elements’ table ?
The table Integron Elements shows all attachment sites (attC, attI) and promoters (Pc, Pi) identified in the predicted integron.
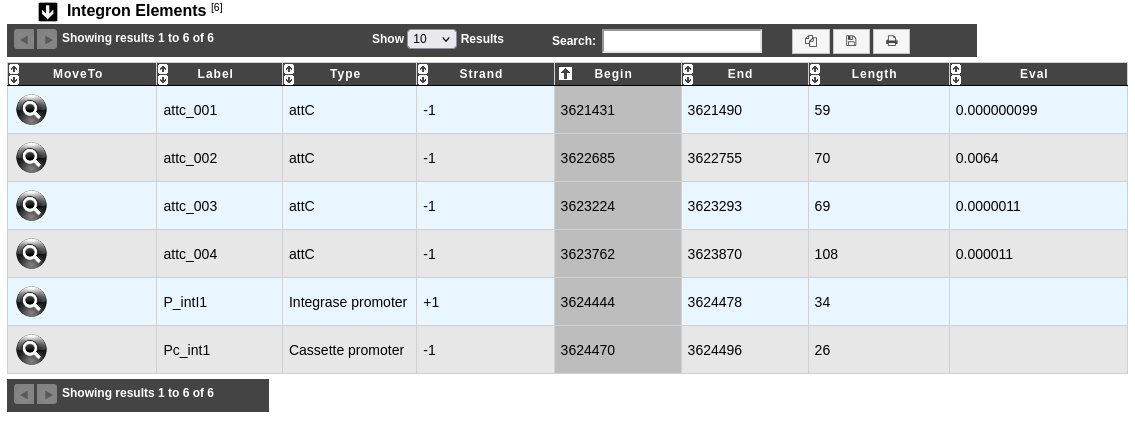
Label: Label of the element.
Type: Type of element (attC, attI, Cassette promoter, Integrase promoter).
Strand, Begin and End: Location of the element on the sequence.
Eval: Evalue of the match between an attC site and the Covariance Model of attC site.
What is the ‘Integron Integrase’ table ?
The table Integron Integrase provides characteristics of the identified integrase intI of the predicted integron.

Label: Label of the genomic object. Click on it allow to access to its annotation page.
Begin, End and Frame: Location of the genomic object on the sequence.
Gene: Gene name if any.
Evidence: automatic/validated.
Type: CDS,fCDS,tRNA,rRNA,misc_RNA,…
Product: Description of the gene product of the genomic object.
Eval: Evalue of the match between the integrase intI and the integrase HMM profil.
What is the ‘Genomic Objects’ table ?
The table Genomic Objects provides informations regarding the genomic objects present in the integron.
You can export the genes by clicking on Export to Gene Cart.
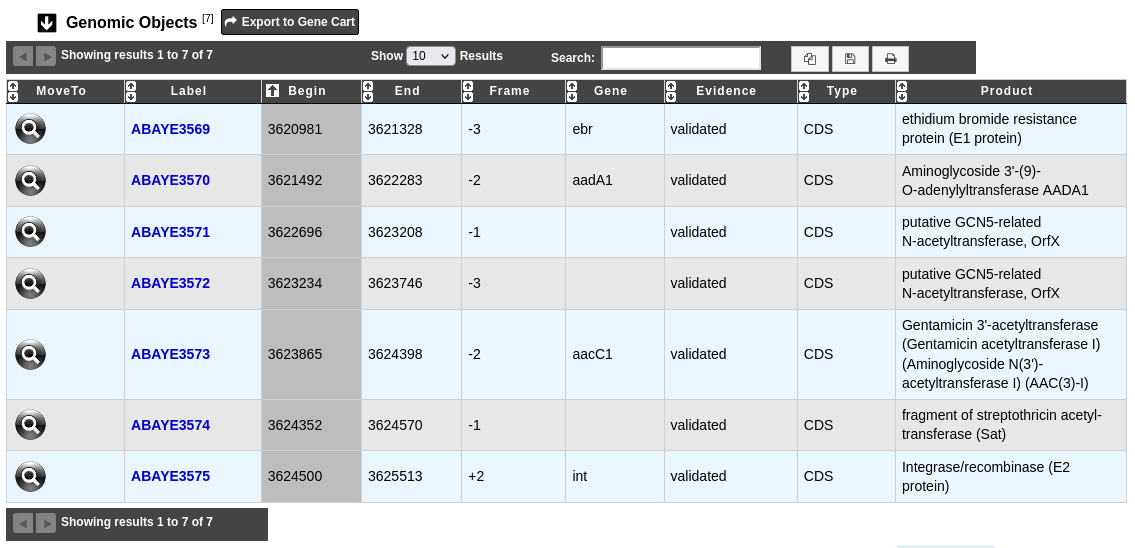
Label: Label of the genomic object. Click on it allow to access to its annotation page.
Begin, End and Frame: Location of the genomic object on the sequence.
Gene: Gene name if any.
Evidence: automatic/validated.
Type: CDS,fCDS,tRNA,rRNA,misc_RNA,…
Product: Description of the gene product of the genomic object.