Pathway Curation¶
How to access to the Pathway Curation Tool?¶
Pathway Curation tool is accessible in the Metabolism section of the main navigation menu.
What is the usefulness of this tool?¶
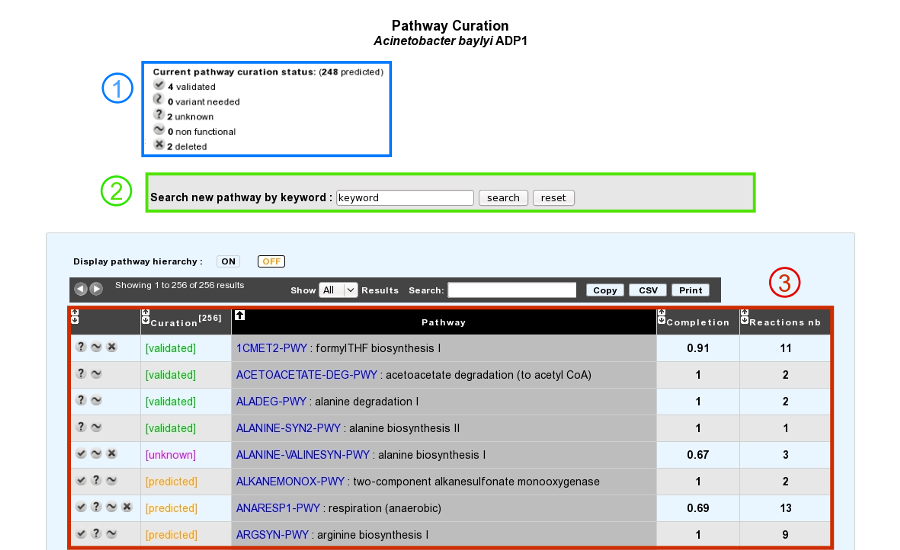
This tool presents a list of predicted MicroCyc pathways in a given organism, coming from pathway-tools software results, for which statuses can be curated by the annotator (3).
The current state of curation is resumed at the top of the page (1).
It is also possible to add a new MetaCyc pathway in the organism if this one is not predicted by the BioCyc pathologic algorithm (2).
How to read the result table?¶
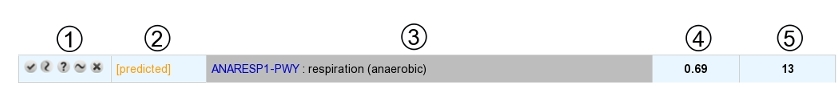
The table is composed of 5 columns:
1 : buttons to change the pathway status (see below for a list of possible statuses)
2 : current curation status of the pathway
3 : pathway identifier and name
4 : completion of the pathway in the organism
5 : number of reactions in the pathway (excluding spontaneous reactions)
Above the table, an option allows users to display or not the MetaCyc hierarchy.
What are the different curation statuses?¶
Users are able to curate the prediction for a given organism by assigning different statuses.
The different statuses are:

predicted: Predicted by the BioCyc pathologic algorithm (default one).
validated: Curated as a functional pathway (all the reactions of the pathway are supposed to exist in the organism).
variant_needed: The predicted pathway is not completely correct for the organism (i.e. some reactions may not be present in the organism but no better pathway definition exists in MetaCyc). Thus, a new pathway variant definition is needed.
unknown: Not enough evidence to declare the pathway as functional (i.e. validated status).
non_functional: The pathway has been lost in the organism and is no more functional (i.e. due to gene loss or pseudogenisation events).
deleted: Curated as a false positive prediction.
A complete pathway cannot be deleted.
How to use this tool?¶
The pathway status can be modified using the buttons “validated”, “variant_needed”, “unknown”, “non_functional” and “deleted”.
Moreover, it is possible to add a MetaCyc pathway which has not been predicted by using a keyword search tool.
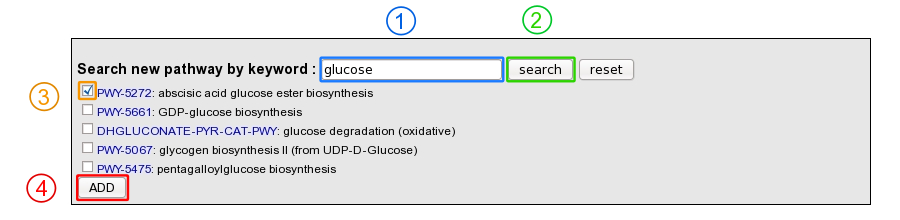
1: Enter a keyword relative to the pathway of interest (ex: glucose).
2: Click on “search” button.
3: Select the correct pathway
4: Click on “Add” button in order to set the pathway as present in the organism.