Integron¶
What are Integrons?¶
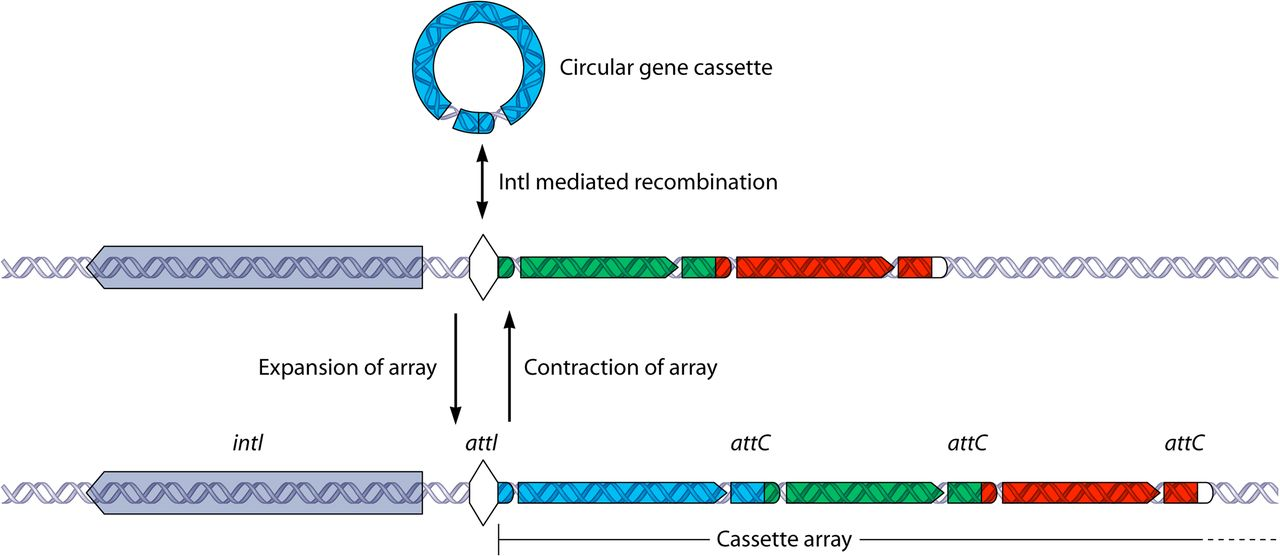
Integrons are versatile gene acquisition systems commonly found in bacterial genomes. They are ancient elements that are a hot spot for genomic complexity, generating phenotypic diversity and shaping adaptive responses. Integrons are composed of three essential core features:
intI : a gene which encodes for an integron integrase whose protein catalyzes recombination between incoming gene cassettes and the second feature, an integron-associated recombination site.
attI : attachment integrase is a proximal recombination site which is recognized by the integrase and at which gene cassettes may be inserted.
Pc: a promoter which directs transcription of a cassette-encoded gene.
Integrons acquire new genes as part of gene cassettes. These are simple structures, usually consisting of a single open reading frame (ORF) bounded by a cassette-associated recombination site known as attC. Circular gene cassettes are integrated by site-specific recombination between attI and attC, a process mediated by the intI. This process is reversible, and cassettes can be excised as free circular DNA elements. Insertion at the attI site allows expression of an incoming cassettte, driven by the adjacent Pc promoter.
Reference:
Gillings MR. 2014. Integrons: past, present, and future. Microbiol Mol Biol Rev 78:257–277.
What is IntegronFinder?¶
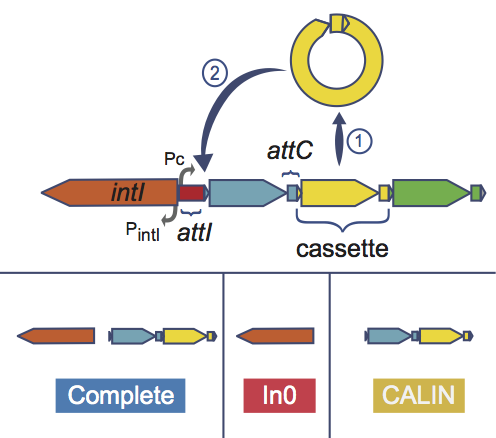
IntegronFinder is a tool that detects integrons in DNA sequences with high accuracy. It is accurate because it combines the use of HMM profiles for the dectection of essential protein, the site-specific integron integrase, and the use of Covariance Models for the detection of the recombination site, the attC site. This tool also annotates gene casettes however we use our own annotations to make it run. IntegronFinder distinguishes 3 types of elements:
Complete integron: integron including an integrase and at least one attC site
In0 element: integron integrase only, without any attC site nearby
CALIN element: The clusters of attC sites lacking integron-integrases (CALIN) are composed of at least two attC sites
Know more about IntegronFinder
How to access to Integrons data ?¶
IntegronFinder predictions are available through the Comparative Genomics section, in the main navigation menu.
What is the ‘Integron clusters’ table?¶
This table enumerates all integron clusters predicted for the selected organism and its replicons.
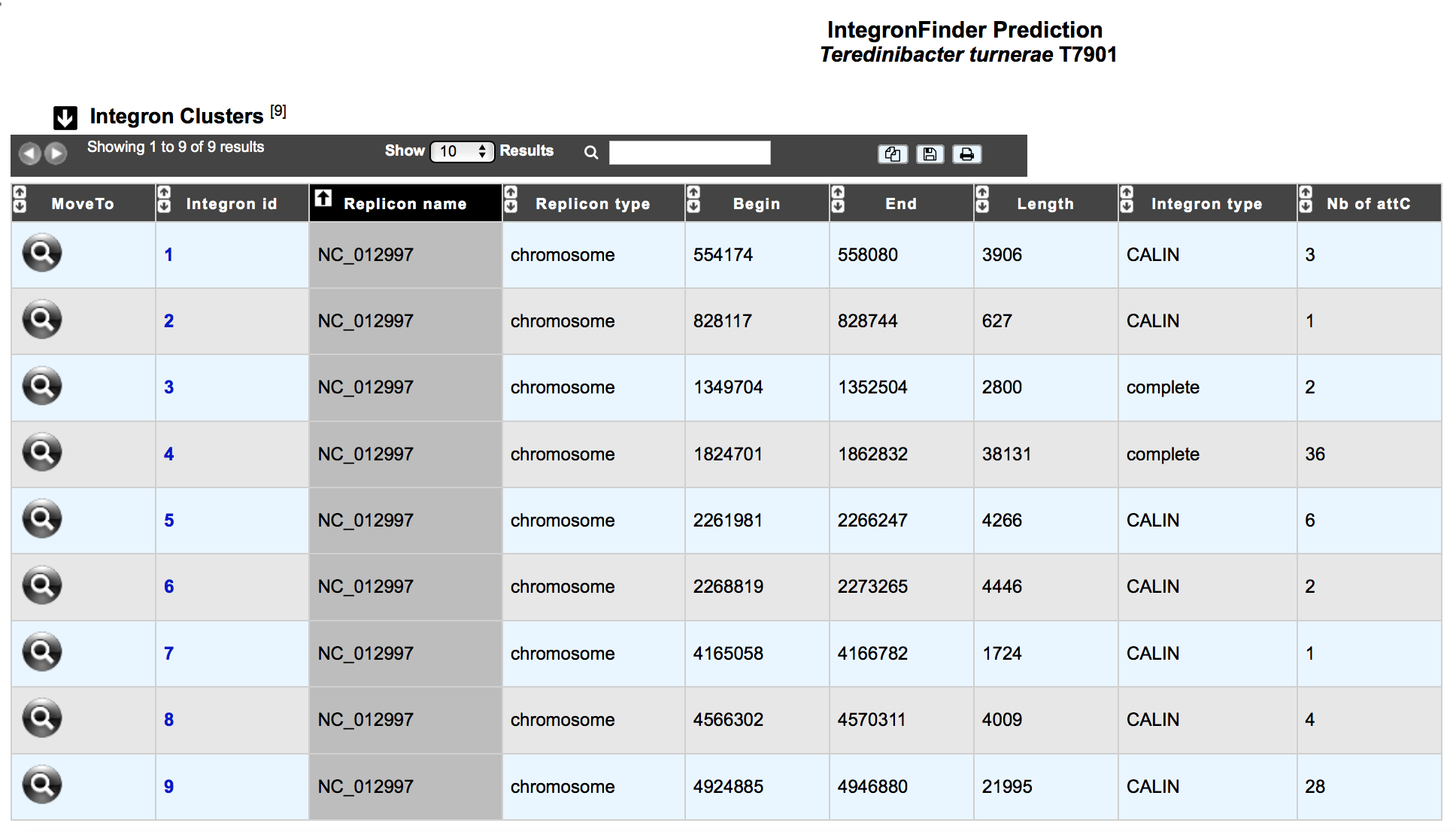
How to explore Integron clusters?¶
The IntegronFinder cluster visualization window can be accessed by clicking on any cluster number in the Integron Id field. This window allows you to access to a detailled description of the integron structure.